Mendelian Randomization (MR) Services
Context
Unlocking Causality from Genomic Data
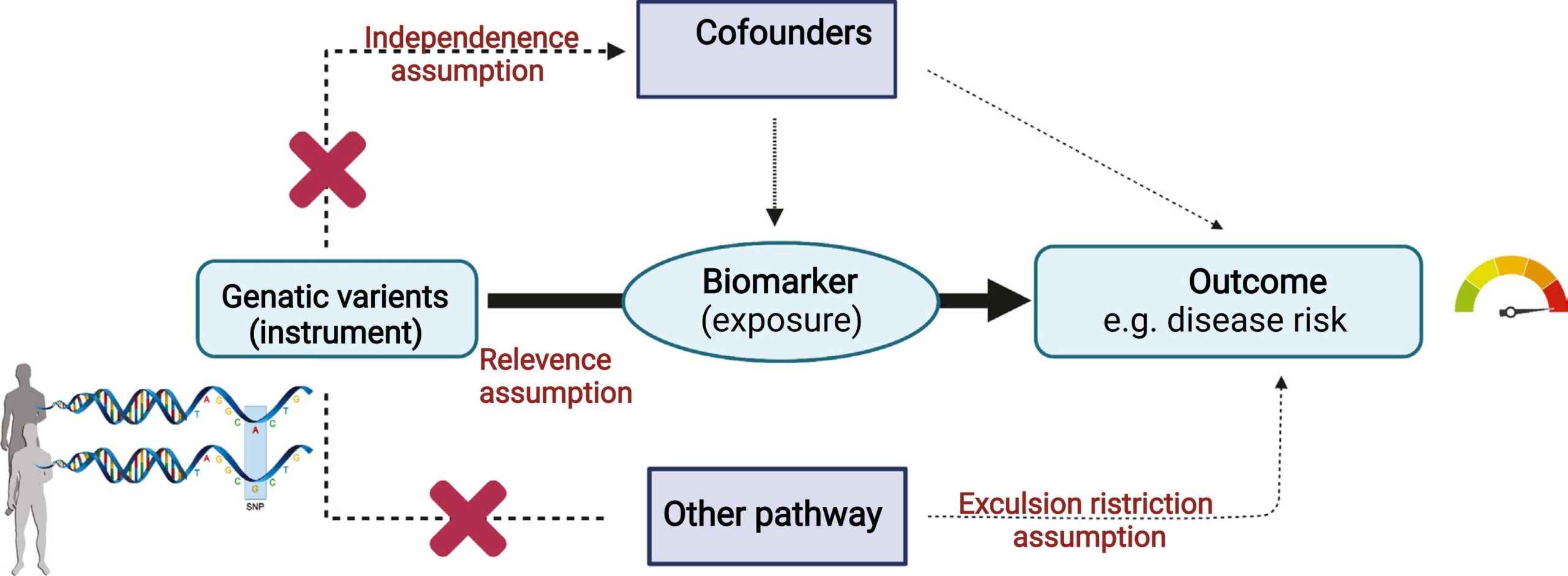
With the rise in large-scale genomic datasets, researchers face challenges in identifying causal relationships between genetic variants and diseases. Traditional observational studies are often confounded by complex interactions, making it challenging to pinpoint causal factors for a disease or trait.
Mendelian Randomization (MR) offers a powerful solution. By leveraging variation in genes with known function, MR empowers researchers like you to examine the ‘unconfounded’ causal effect estimate of a modifiable exposure to disease in observational studies.
The growing utilization of high-throughput proteomics platforms in large-scale genotyped biobanks (e.g., UKBB-PPP) provides new opportunities to obtain biological insights from GWAS data. Exploiting MR approaches can identify potentially causal effects of circulating proteins on diseases/ traits. MR helps in identifying disease risk factors, and drug targets, providing early predictions of ongoing trials, predicting adverse events, and also aiding in drug repurposing.
How We Can Help
At Aganitha, we recognize that implementing MR involves navigating intricate statistical methodologies and the complexities of handling genomic data at scale. Our cross-functional team of AI/ML experts, bioinformaticians, and statistical geneticists is here to support you at every stage.
Customizable MR Pipelines
We leverage the power of Mendelian Randomization to address a wide range of scientific questions, tailoring our pipelines to your specific needs. Whether you’re utilizing one-sample, two-sample, or bidirectional MR designs to investigate causal relationships between exposures and outcomes, we provide the expertise and tools to support your research.
Robust Statistical Analysis at Scale:
Our expertise encompasses a range of robust statistical methods to address the multifaceted nature of MR analysis, even with large-scale datasets. We control for horizontal pleiotropy, confounding variables, and instrument variable bias, ensuring the validity of your causal inferences. We also employ advanced techniques to tackle complexities such as single-locus colocalization, multi-locus heterogeneity assessment, and cis-MR or functional enrichment of IVs.
Clear and Actionable Insights
We provide easy-to-interpret visualizations highlighting key findings, potential causal pathways, and implications for your research.
Our Pipeline
We follow a structured, efficient process to ensure your MR studies deliver precise, actionable results
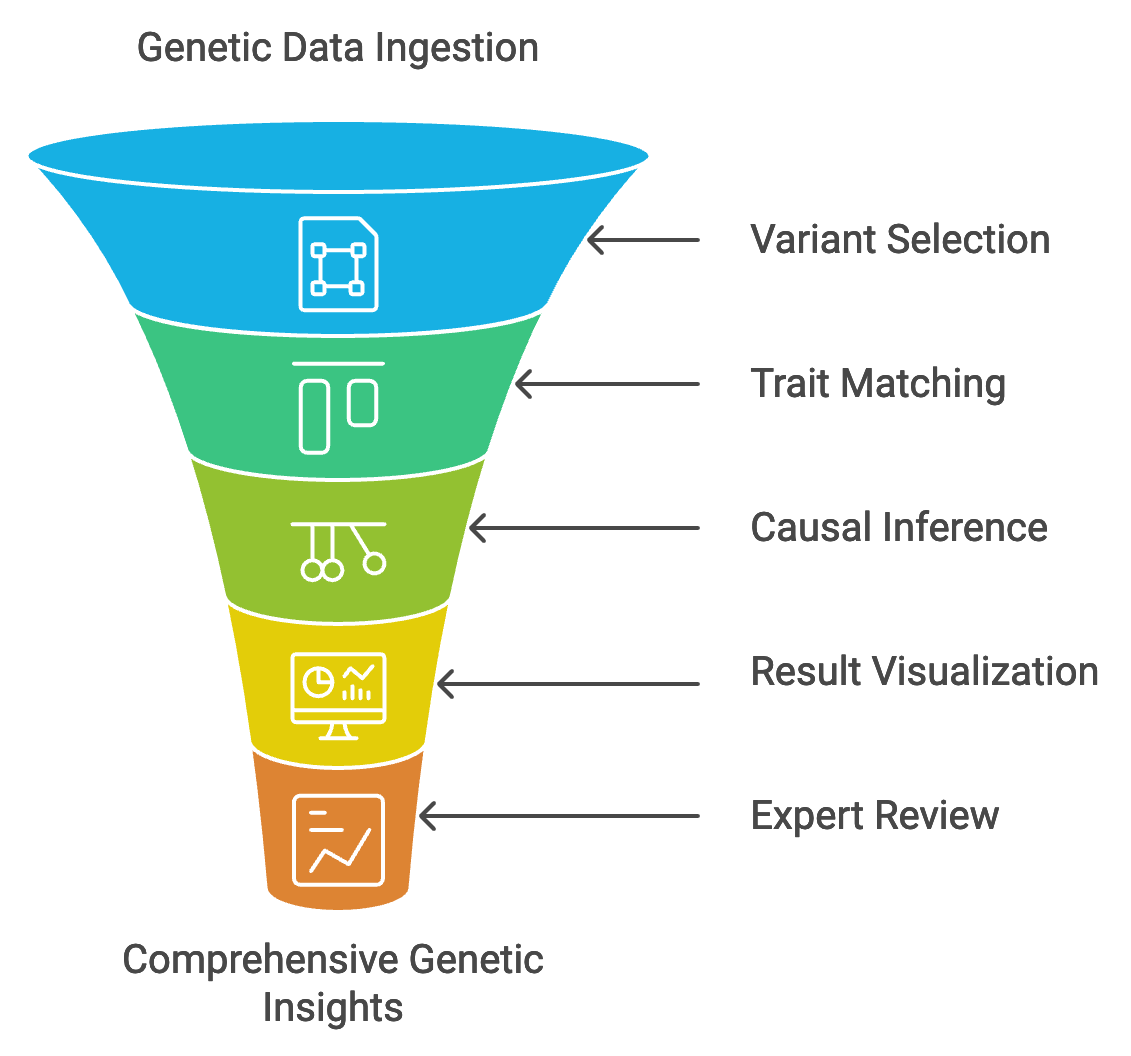
- Data Ingestion: We start by ingesting your genetic data from biobanks or sequencing efforts, whether you’re working with whole-genome or exome sequencing.
- Genetic Variant Selection: We assist in selecting or filtering genetic variants that serve as instrumental variables, ensuring robust causal inference.
- Trait and Outcome Matching: Our platform enables the matching of genetic variants to traits and outcomes of interest based on your study design (one-sample or two-sample MR).
- Causal Inference and Statistical Analysis: Using customized MR scripts, we perform instrumental variable analyses to assess the traits’ causal effects on the outcomes, generating relevant statistical outputs.
- Result Visualization: We provide clear and comprehensive visualizations, including scatter plots, forest plots, funnel plots, and exposure vs. trait heatmaps (for MVMR), to facilitate your understanding of the causal relationships and potential confounders identified in your MR analysis.
- Review and Interpretation: Our team of experts goes beyond visualization, offering in-depth interpretation of your results, drawing upon their extensive knowledge of MR methodologies and statistical techniques, including one-sample and two-sample MR, univariate and multivariate MR, IV selection and bias assessment, heterogeneity analysis (MR-Egger), sensitivity analysis, and subgroup analysis.
Highlights
Key components & strengths
Addressing Complexities:
We adeptly handle single locus-colocalization, multi-locus heterogeneity assessment, and cis-MR or functional enrichment of IVs.
Accounting for Confounding Factors
We employ methods to account for confounding variables like age, sex, and population stratification, enhancing the reliability of your results
Managing Pleiotropy
Our sensitivity analyses address the possibility of genetic variants influencing multiple traits, ensuring robust causal inference.
Handling Large-Scale Datasets
Our computational infrastructure is designed to handle the demands of modern genomics, enabling MR on even the most extensive datasets.
Discover our offerings across the biopharma value chain
Our Solutions
Customizable platforms and solutions across the drug discovery value chain from target discovery to therapeutic development
Read More
Our Services
Offering services in computational sciences and technology to complement biopharma R&D
Case Studies
Helping clients accelerate drug discovery and development by building computational biology, computational chemistry, technology, and cloud solutions
Read More