Sequence Structure Co-Modeling
The designable antibody space is too large and it is practically impossible to evaluate all possible combinations to come up with an antibody. Generative AI provides us with the capability to evaluate this unexplored space to come up with potential new sequences for antibodies. However, going by just the sequence of an antibody does not provide insights into binding to the antigen. Structural consideration along with sequence would help solve this challenge by selecting the antibodies with structures that would bind to the target antigen.
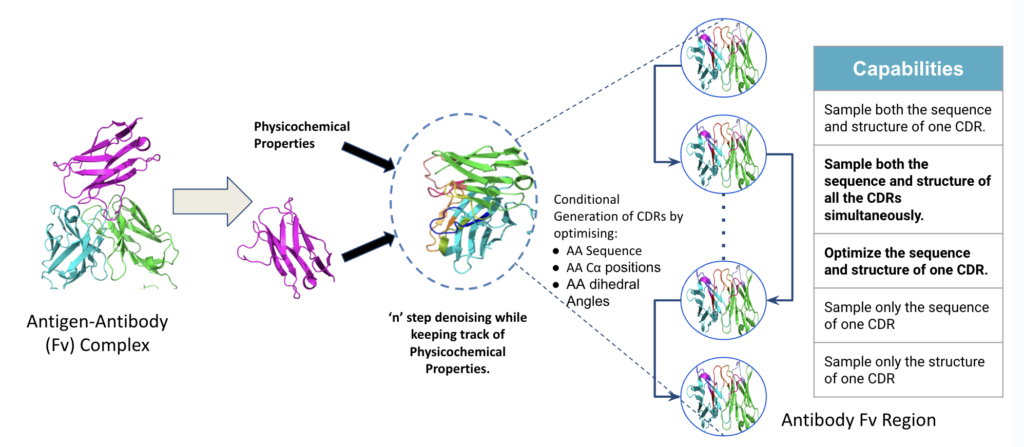
Generative AI can be used to explore this combinatorial space while considering sequence and structural co-modeling to design antibodies to a target antigen of interest. Co-modeling of antibodies is done using machine learning (ML) models which jointly model the sequence and structure of a set of known antibodies. The model is trained on the distribution of antibody sequences and structures by taking into account the relation between them. The model learns to represent the probability distribution of antibody sequences and structures, taking into account the relationship between the two. These models are used to conditionally design antibodies.
Aganitha’s Sequence-Structure co-design framework, as visualized above, presents an advanced solution to the challenges of antibody design. It’s a deep generative model that cohesively brings together sequences and structures of CDRs, using diffusion probabilistic models and equivariant neural networks.
The conditional generation process ensures a harmonious blend of sequence, structure and developability properties, producing antibodies that are not only theoretically sound but also positioned for success for high efficacy in real-world applications.
We have the team and computational capabilities (research scientists, AI engineers, AI models, software tools) that can work with you on your antibody engineering project.